Troubleshooting
This page offers some useful tips to troubleshoot based off some of the problems we encountered when building the simulator.
Accessing Variables in the Simulator
In order to troubleshoot a script utilizing the simulator, you may want to access different variables that are found in the simulator, but not locally available in your script. This can help with troubleshooting issues by making sure everything is defined as you think it should be.
A detailed description of the matrix setup of the model variables can be found at Detailed Model Description.
Note:
Only varibles that have the “self” attachment are available to be accessed from outside the simulator (Ex: self.variable_name).
If you wish to pull out a variable that does not currently have the self attachment, simply add “self.” before every occurence of that variable in the simulator.
In order to access any variable with the “self” attachment (Example Below):
The bulk of the simulator’s code (excluding the rate equations function) exists in the RSDs_sim() Python class. Therefore, as you would to run a simulation, you must first instantiate an object of the RSD_sim class. Details to do this can be found here.
Assuming the model is downloaded, imported, and instantiated accessing a variable is very similar to calling one of the functions. You simply call the variable name with the name of the class object’s attachment. For example, if you defined the model instantiation as “model”, then the format for accessing a variable would be “model.variable_name”.
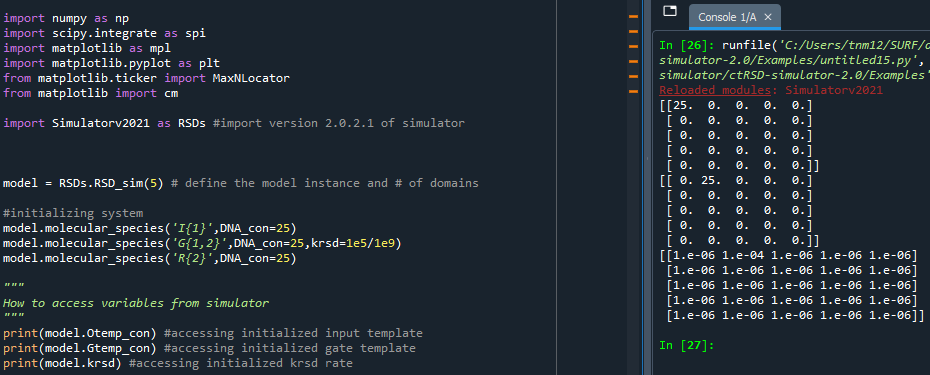
How to access variables present in simulator
Critical values to check for customized simulations include:
- The matrices holding DNA template concentrations
These are specified as model.[name]temp_con, where [name] can be G, O, TG, F, AG, CG
- The matrices holding initial concentrations if these were changed in molecular_species()
These are specified as model.[name]_ic, where [name] can be uG, G, GO, O, TG, F, AG, CG, R, S, Q, AGOa, AGOb, AGFb, CGOa, CGOb
- The matrices holding rate constants if these were changed in global_rate_constants() or molecular_species()
These are specified as model.[name], where [name] is the name of the rate constant (ktxnO, krsd, krz, kdrd, kssdO, etc)
Not specifying enough domains in RSD_sim() initialization
When initializing the model with RSD_sim(), the default number of domains used to initialize the model is 5. If the highest index of any domain of a species you define in molecular_species() is greater than 5 then an “index out of bounds” error will appear. For example:
# create the model instance
model = RSDs.RSD_sim() # default # of domains (5 domains)
# specify species involved in the system
model.molecular_species('G{7,2}',DNA_con=25)
will produce an error because index 7 of G{7,2} exceeds the default number of domains (5) specified with RSD_sim():
line 30, in <module>
model.molecular_species('G{7,2}',DNA_con=25)
File "filepath", line 779, in molecular_species
self.Gtemp_con[gateInd1,gateInd2]=DNA_con
IndexError: index 6 is out of bounds for axis 0 with size 5
To fix this, the total number of domains initialized in RSD_sim() needs to be at least 7:
# create the model instance
model = RSDs.RSD_sim(7) # increasing total number of domains for initialization to 7
# specify species involved in the system
model.molecular_species('G{7,2}',DNA_con=25)
Simulations taking a long time
The simulation time increases as the number of domains in the system and the simulated time increases. So to speed up specific simulations you can try to minimize the number of domains necessary to describe the system and specify this minimum number of domains in RSD_sim() during initialization. You can also decrease the total simulated time (t_sim).
We primarily saw issues with simulations taking a long time to solve for systems with comparator gates (CG) and additional tips for those specific types or simulations are covered below.
Changing the smethod in simulate can also speed up simulations but numerical instabilities are also possible.
Speeding Up Comparator Gate Simulations
Due to comparator gate reactions utilizing very disparate time scales, simulations using comparator gates may have a much slower time efficiency than the other reactions. Below are some different options for reducing the simulation time when using comparator gates.
- Troubleshooting options:
Utilize the BDF solving method. BDF is an implicit method meant for stiff equations, so it can help to process the CG reactions quicker. The solving method used can be changed in simulate.
If you are looping through the simulator many times, parallelizing these loops can also greatly speed time efficiency. Python offers many different ways to create parallel loops, such as the multiprocessing package.
Overriding Rate Constants
Since both molecular_species and global_rate_constants offer the ability to change the rate constants available in the simulator, you may find one function overriding changes you made in the other.
- Troubleshooting Tips (Examples Below):
Both functions will only alter rate constants that are specified in the call of the function. If you do not specify a particular rate constant, then neither functions will change it from its most recent value.
The order of the changes made by the two functions depends on the order in which the functions are called in the script. Whichever function is called first will have its changes come into effect first. Therefore, if you call both functions for the same rate constant, or either functions multiple times for the same rate constant, then the last function call will reflect the most recent value of that rate constant.
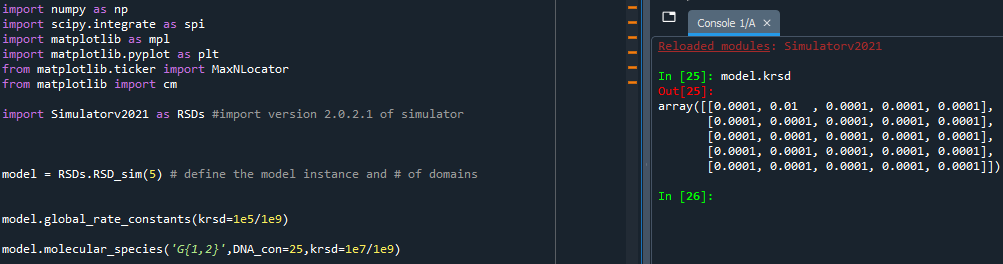
global_rate_constants before molecular species
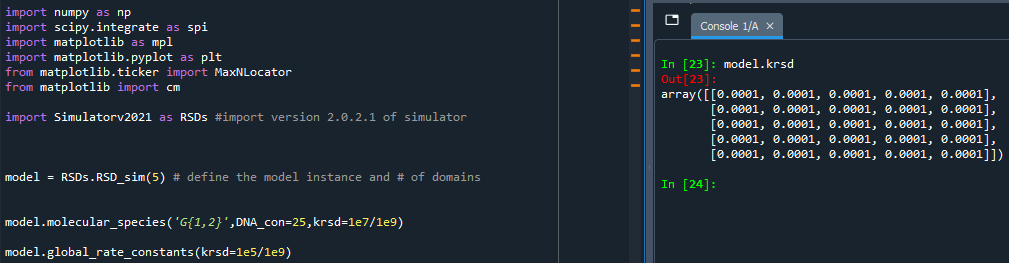
molecular_species before global_rate_constants