Model Reactions and Implementation
Model overview
The reactions currently incorporated in the model are shown below. The simulator converts all the reactions below into the appropriate rate equations derived from mass action kinetics. This produces a system of ordinary differential equations that is then numerically integrated with the solve_ivp() function in Python’s SciPy package. Concentrations in the simulator are in nM and time is in seconds. The default values for rate constants can be found here
The curly bracket notation specifies the nomenclature of the species where i,j,k,…etc represent the domain numbers with the first index specifying the input domain and the second index specifying the output domain {input,output}. The names of species and the rate constants in the schematics below match how they are defined as variables in the model. The model is matrixed based so the index in the curly brackets also corresponds to the matrix index where the species concentrations are stored, or where the value of individual rate constants are stored in the rate constant matrices. See here for more details on matrix definitions.
The model uses a universal toehold for all strand displacement reactions. So from the model’s perspective all outputs have the same toehold and any output with an output domain index that matches the input domain index of a gate can react with that gate, i.e., O{3,1} will react with G{1,4} and G{1,5}. If it is desired to model a system in which multiple toeholds with different strand displacement rates, the appropriate index of the krsd rate constant matrix can be changed in molecular_species for each gate.
The model treats input species as outputs with identical input-output indices, i.e., I{1} = O{1,1}, I{4} = O{4,4}. The user can specify input species in the model functions, such as when defining species in molecular_species() or when pulling concentrations for analysis with output_concentration(), but internally these will be handled in the output matrices. This implementation means other species that produce outputs with identical input-output indices, such as G{1,1}, AG{3.2,2} should not be used in the model as their outputs will behave as inputs rather than outputs.
There are a few things to note about the way the rate constant matrices are defined and implemented.
For RNA strand displacement reactions, unless otherwise stated, the forward rate constants are associated with the gates. This means the rate constant for a forward strand displacement reaction for a specific gate, for example G{1,3}, can be changed and that rate constant will be used with all outputs that react with that gate, i.e., O{4,1} and O{5,1} will both use the forward rate constant associated with G{1,3}. It is not possible to give O{4,1} and O{5,1} unique forward rate constants for a strand displacement reaction with G{1,3} (or any gate with input domain index 1). Changes to forward RNA strand displacement reactions for individual gates should be done in the molecular_species() function when defining the gates. Changing the forward RNA strand displacement rate constant is not a valid option for outputs in molecular_species().
The reverse rate constants for RNA strand displacement reactions are associated with the outputs. This means the rate constant for a reverse strand displacement reaction for a specific output, for example O{1,3}, can be changed and that rate constant will be used with all GO{} species that react with the output, i.e., GO{4,1} and GO{5,1} will both use the reverse rate constant associated with O{1,3}. It is not possible to give GO{4,1} and GO{5,1} unique reverse rate constants for a strand displacement reaction with O{1,3} (or any output with input domain index 1). Changes to reverse RNA strand displacement reactions for individual outputs should be done in the molecular_species() function when defining the outputs, or when defining the gates – the reverse rate that is defined with a gate with correspond to the output of that gate. NOTE: because inputs are stored on the diagonal of the output matrix the diagonal of the reverse RNA strand displacement rate constant matrix is set to 0. This is because inputs only possess an output domain index and cannot participate in reverse RNA strand displacement reactions
ctRSD Reaction Schematics
Below the subscript “temp” indicates a DNA transcription template that encodes for the RNA species specified in its name.
We model RNA transcription and degradation using a first order approximation for enzyme kinetics. From this approximation the transcription rate is linearly proportional to the template concentration, e.g., ktxnO{k,i}*[Otemp{k,i}] where ktxn{k,i} is the apparent first order rate constant for production of RNA from Otemp{k,i}. Likewise, the degradation rates (if included in the model) are linearly proportional to the concentration of RNA, e.g., kssdO{i,j}*[O{i,j}].
Base ctRSD Reactions
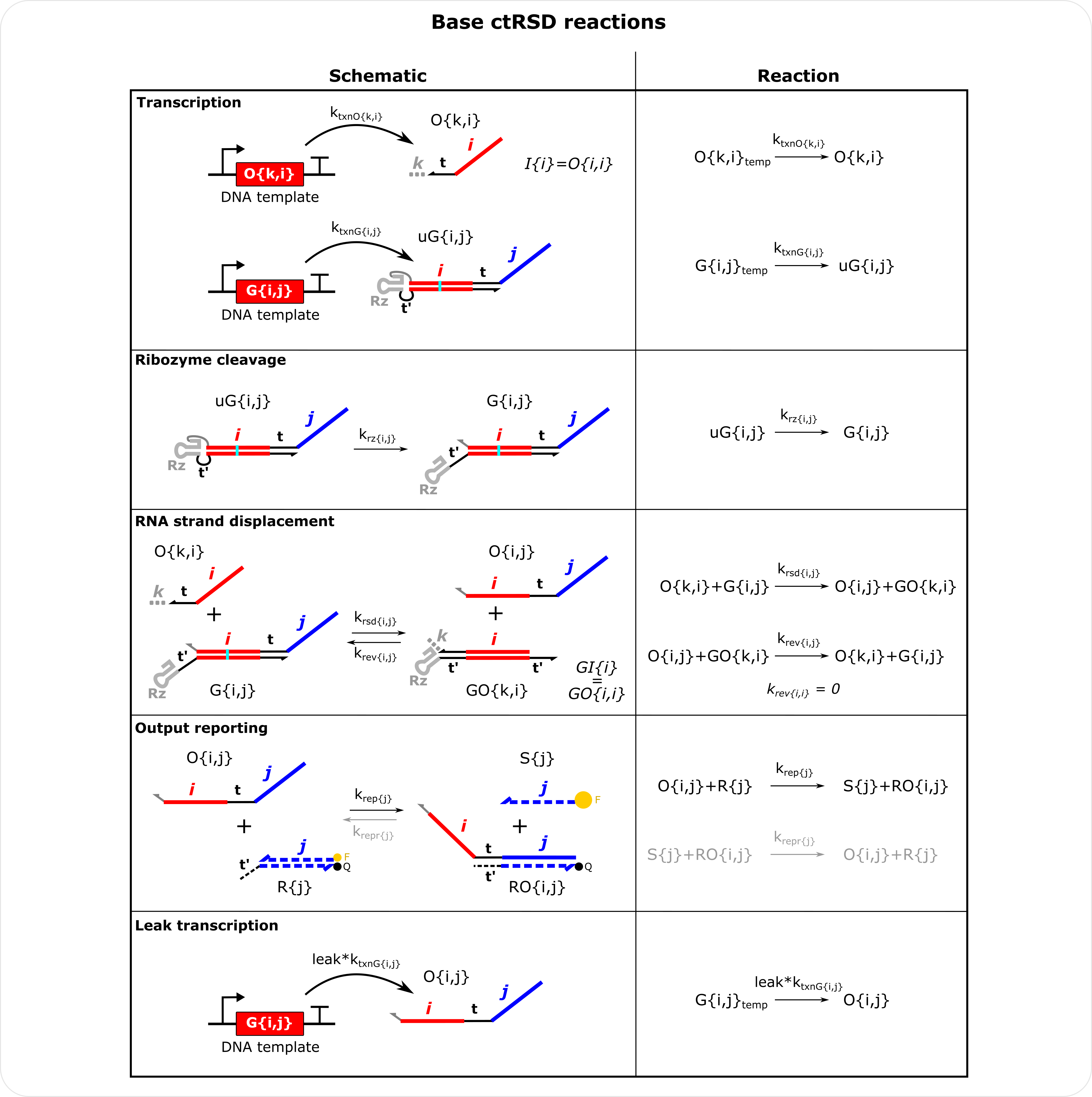
Base ctRSD Reactions Schematics i,j,k represent input domain and output domain indices within the matrices and vectors that make up the model. They can be any integer value from 1 to N, where N is the total number of domains defined in model initialization. In the model inputs are not a separate species. Instead, inputs are modeled as outputs with the same input and output domain (tracked on the diagonal of the output matrix). For example, I{1} is modeled as O{1,1}. The user can still specify inputs in molecular_species() and output_concentration() and the simulator will convert these inputs to the appropriate output. The reverse strand displacement reaction for reporters is shown in gray because the krepr rate constants are initialized as 0 but they can be changed in global_rate_constants() or molecular_species() to simulate reversible reporting reactions. For the leak transcription reaction leak is a percentage (default 0.03). Note the krev rate constant for inputs is 0 as they only have an output domain and cannot reverse a strand displacement reaction. The production of species S{j} can be compared to experiments with DNA reporters.
Fuel Reactions
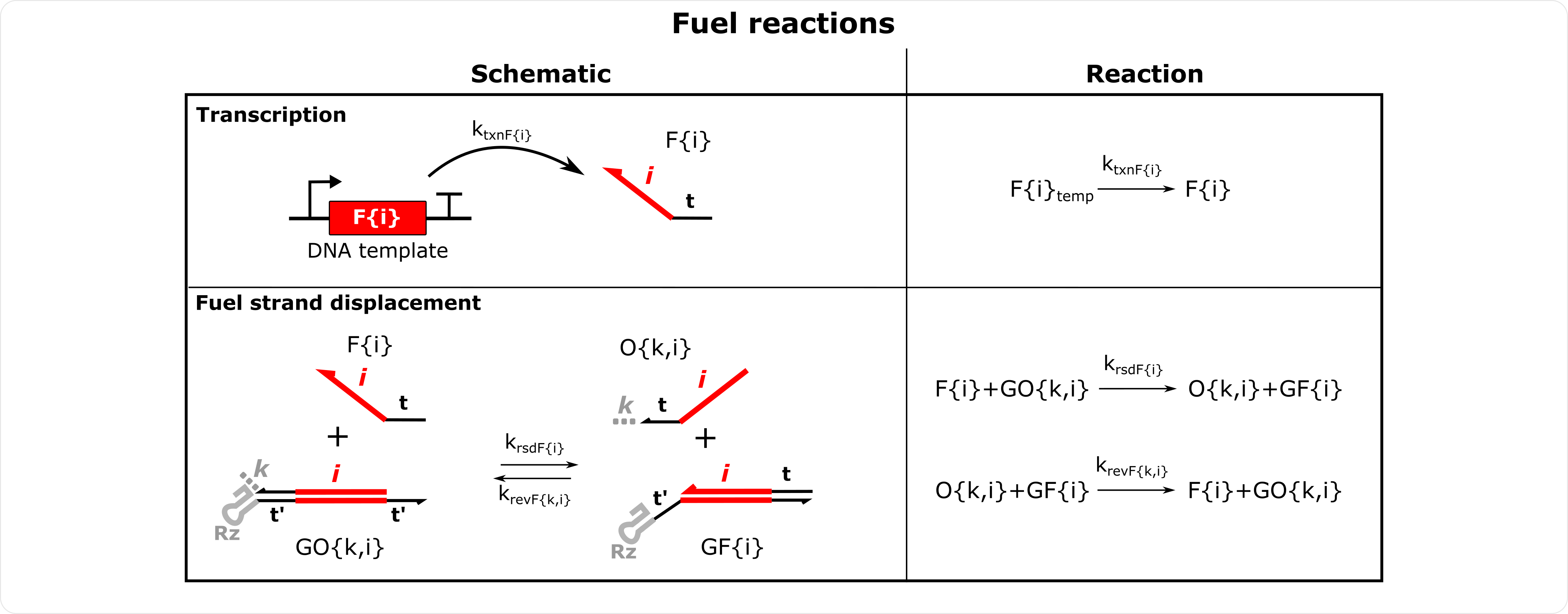
Fuel Reactions Schematics Note the forward fuel strand displacement rate constant is associate with the fuel strand. Fuel reactions with AGs are shown below and are defined similarly.
AND Gate Reactions
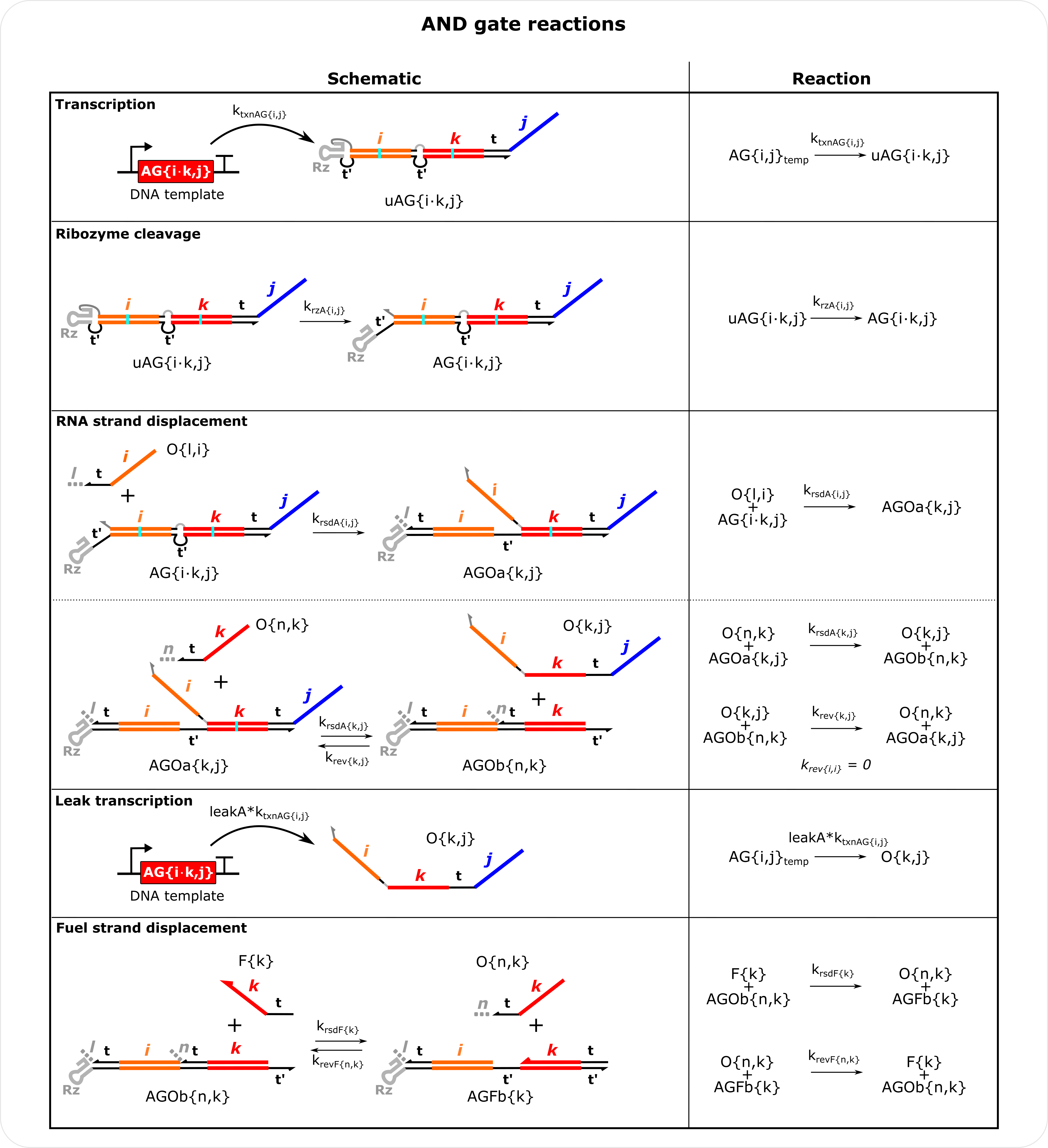
AND Gate Reactions Schematics To reduce the number of species that needed to be tracked a few simiplications were made: The reaction of AG with the first input is considered irreversible. The final output of AG is defined by the second input domain and the output domain so this output will be lumped with outputs from single input gates that have the same indices i.e., in the model O{k,j} from AG{i.k,j} is the same as O{k,j} from G{k,j}. Note there is not a unique reverse strand displacement rate constant for outputs from AG. The same matrix used for outputs from a single input gate is used. Fuel reactions with the first input domain of AGs is not considered, but in experiments such a reaction could occur. As with G, the leak transcription reaction leakA is a percentage (default 0.06). Note AGOa is defined by the second input domain and the output domain of AG while AGOb is defined by the indices of the second output bound to the gate.
Degradation Reactions
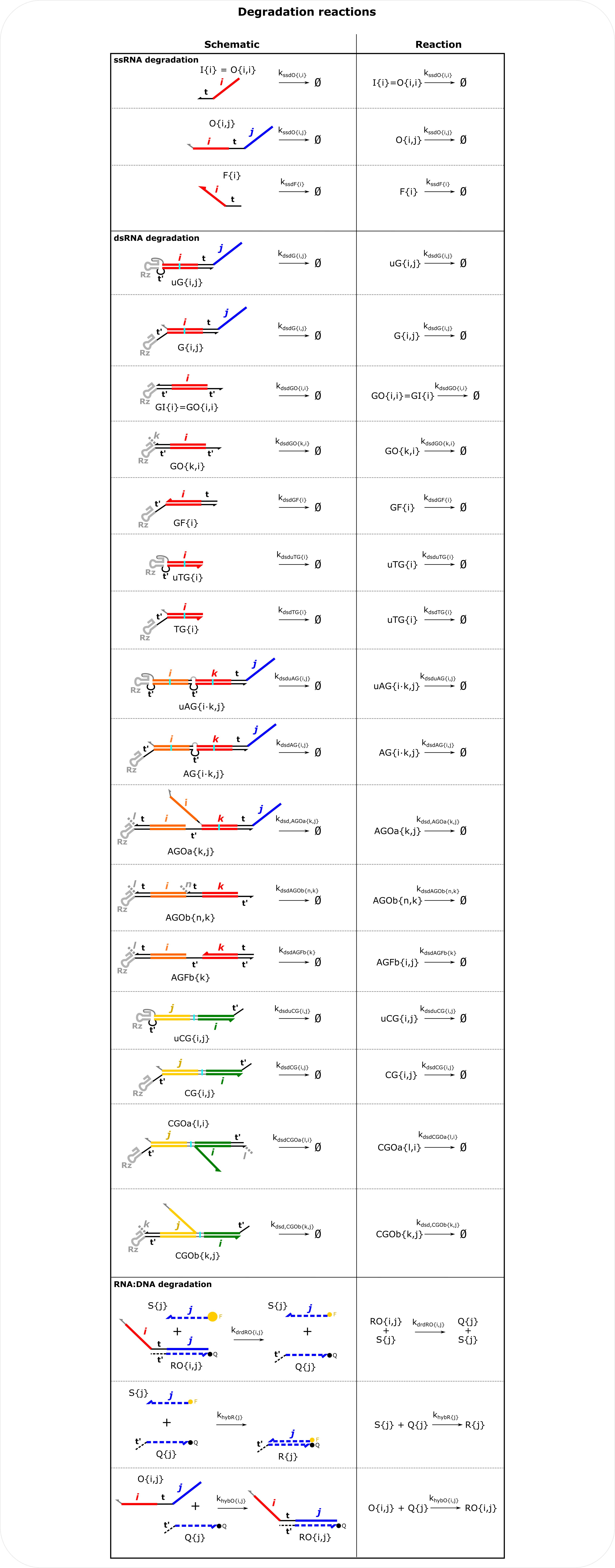
Degradation Reactions Schematics Inputs (I) and GI species are shown separately here because they can be specified in molecular_species() and output_concentration() but they are actually modeled as O and GO, respectively, with identical input and output domain indices.
Thresholding Reactions
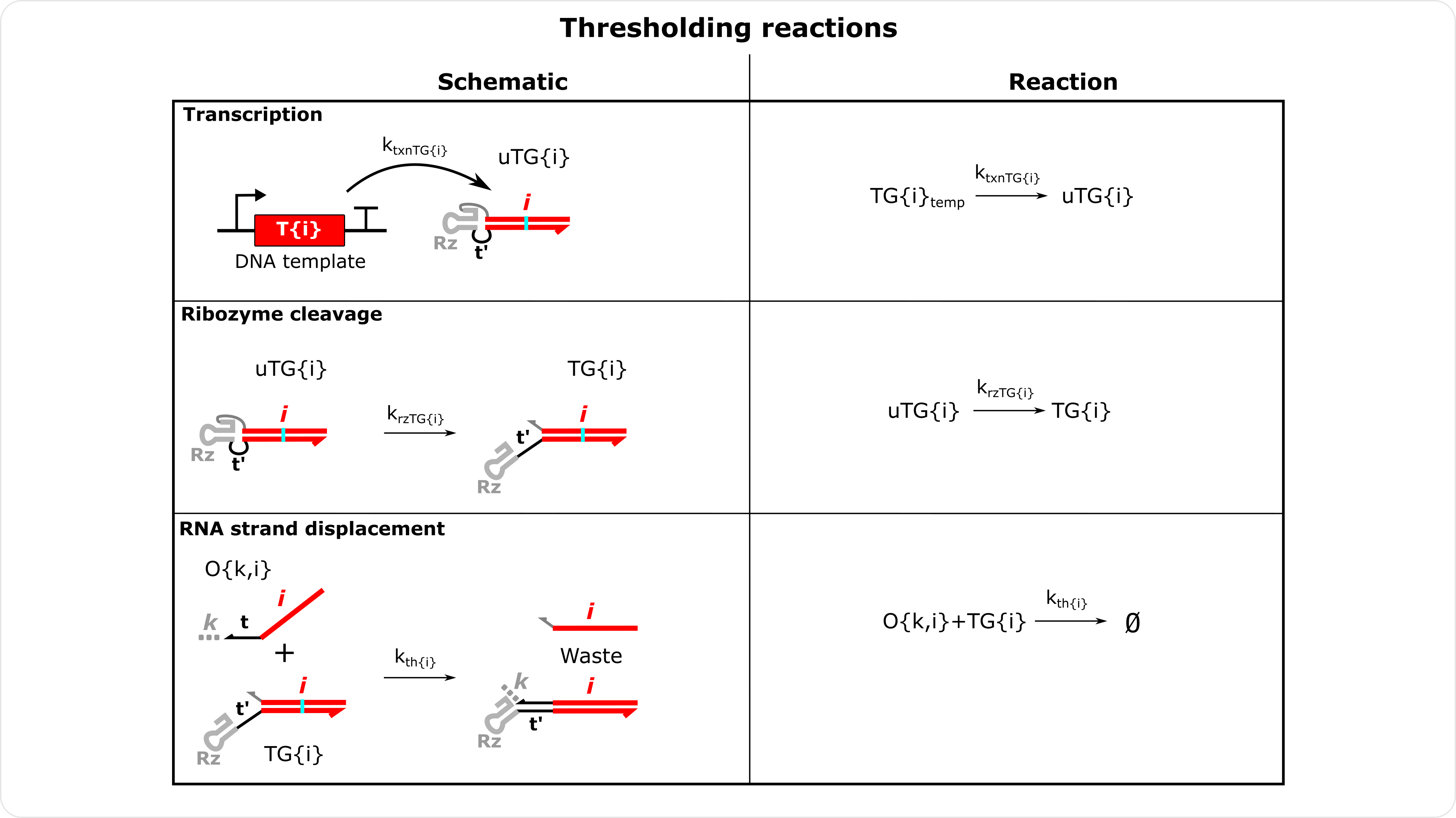
Thresholding Reactions Schematics These reactions represent irreversible reactions without an output, essentially a sink for specific signals in a circuit. So these gates only have an input index.
Comparator Gate Reactions
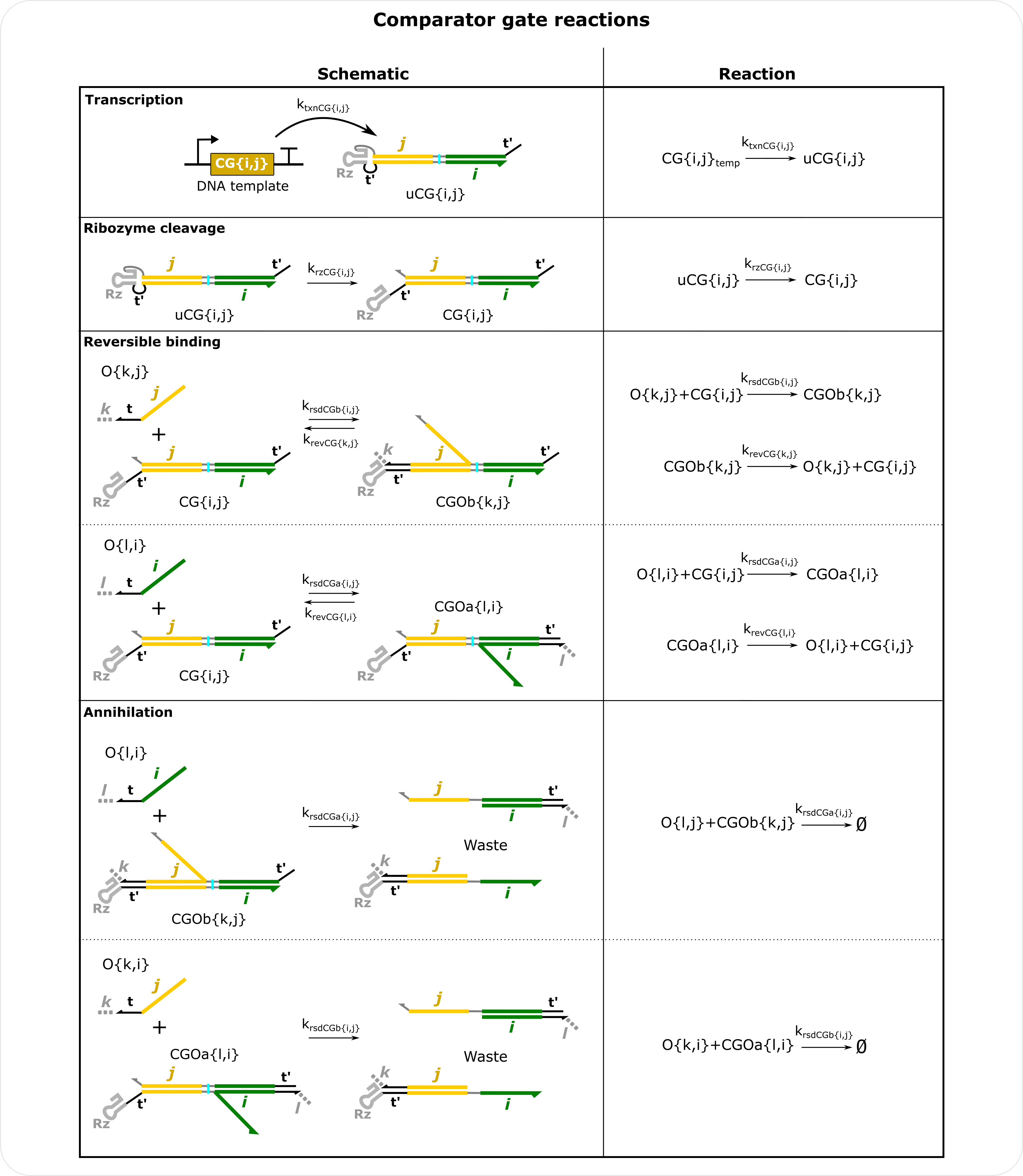
Comparator Gate Reactions Schematics There are two forward strand displacement rate constants for each CG, krsdCGa that corresponds to reactions with the i domain and krsdCGb that corresponds to reactions with the j domain. Note the reverse strand displacement rate constants still follow the outputs, e.g., the rate that O{k,j} dissociates from CG is defined by O{k,j} and not by the j domain of CG. This means outputs with the same output domain but different input domains can have different krevCG values. molecular_species() has an option to change the reverse reaction rate for all outputs with the same output domains when defining CG (krevCGa and krevCGb). Note CGOa and CGOb complexes are defined by the indices of the outputs bound to the comparator gate. For simulations containing more than one CG, the same input domain cannot be repeated in the same index for two gates. For example, CG{i,j} and CG{k,j} will result in an incorrect result because both gates have domain j in the second index. This should be changed to CG{i,j} and CG{j,k} so that domain k is in a different index for the two gates. See the Three comparator gate example.
Detailed model description
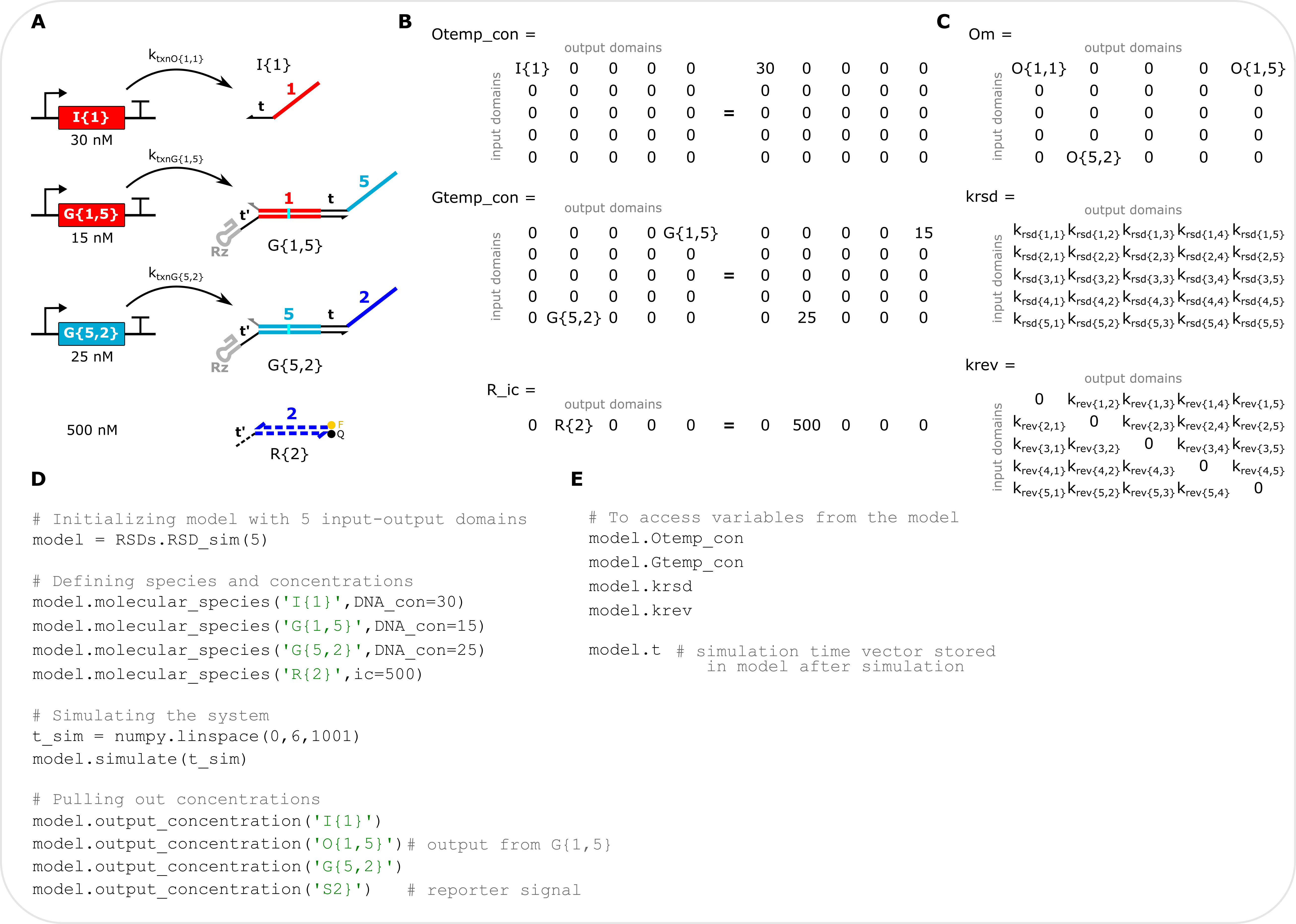
Example with selected matrix descriptions (A) Schematic of a specific system to simulate, composed of and input, two cascaded gates, and a DNA reporter. Concentrations correspond to the DNA templates and reporter. (B) Matrix representation in the model of the defined system. Each column of a matrix corresponds to a specific output domain and each row corresponds to a specific input domain. Left: variables of the defined species with indexes shown. Right: variables with the defined concentrations shown. Otemp_con stores the concentrations of output (and input) DNA templates. Gtemp_con stores the concentrations of gate DNA templates. R_ic stores the initial concentration of DNA reporters. (C) Matrix representation of the output matrix (Om) and two rate constant matrices. Note Om stores both the outputs from the two gates in the system as well as the input because inputs are modeled as outputs with the same input and output domain. The krsd and krev matrices hold all the forward and reverse RNA strand displacement rate constants for all possible gates and outputs, respectively. The diagonal of the krev matrix, where the inputs are stored, is set to 0 because inputs cannot reverse RNA strand displacement reactions given that they only have output domains. The values of every entry in these matrices can be changed simultaneously with global_rate_constants() or the values of individual entries can be changed by specifying a new rate constant value for a specific species in molecular_species(). (D) Example code for setting up and simulating the system in panel A. (E). Commands to access the variables in panels B and C, see Troubleshooting.