Sequence Compiler
Sequence compiler is a function avaliable in the simulator that takes in a species name as an input (same nomenclature used elsewhere in the simulator), among other function inputs, and outputs the DNA sequence that encodes for the RNA species that meets the input specifications.
Note:
Although it is a function within the simulator, and is accessed in the same way as the other functions, it does not require a system to be simulated to be used. Hence why its documentation is being kept seperate from the other functions. It just requires that the simulator package be imported.
Sequence compiler was included in the simulator so that users could have all the useful tools in designing ctRSD circuits in one place.
Download Domains List
Below is a link to download a provided spreadsheet that contains the list of domains for ctRSD reactions used to compile the sequences. The local file path on where this file is stored is an input to the sequence compiler function. Many of the optional inputs to the sequence compiler use the names of sequence domains contained in this Excel workbook - they can be accessed by specifying the name in the first column of the Excel workbook with the appropriate optional parameter. To add custom sequences for a domain, one can add a sequence domain with a unique name to a local copy of the Excel workbook in the correct sheet and then use the name of that sequence domain in the sequence compiler optional inputs.
Function Overview
The figures below illustrate the various options for the sequence compiler. Note, the comparator gates (CG) and thresholding gates (TG) have not been exhaustively tested in experiments and their exact designs might be subject to change in future iterations.
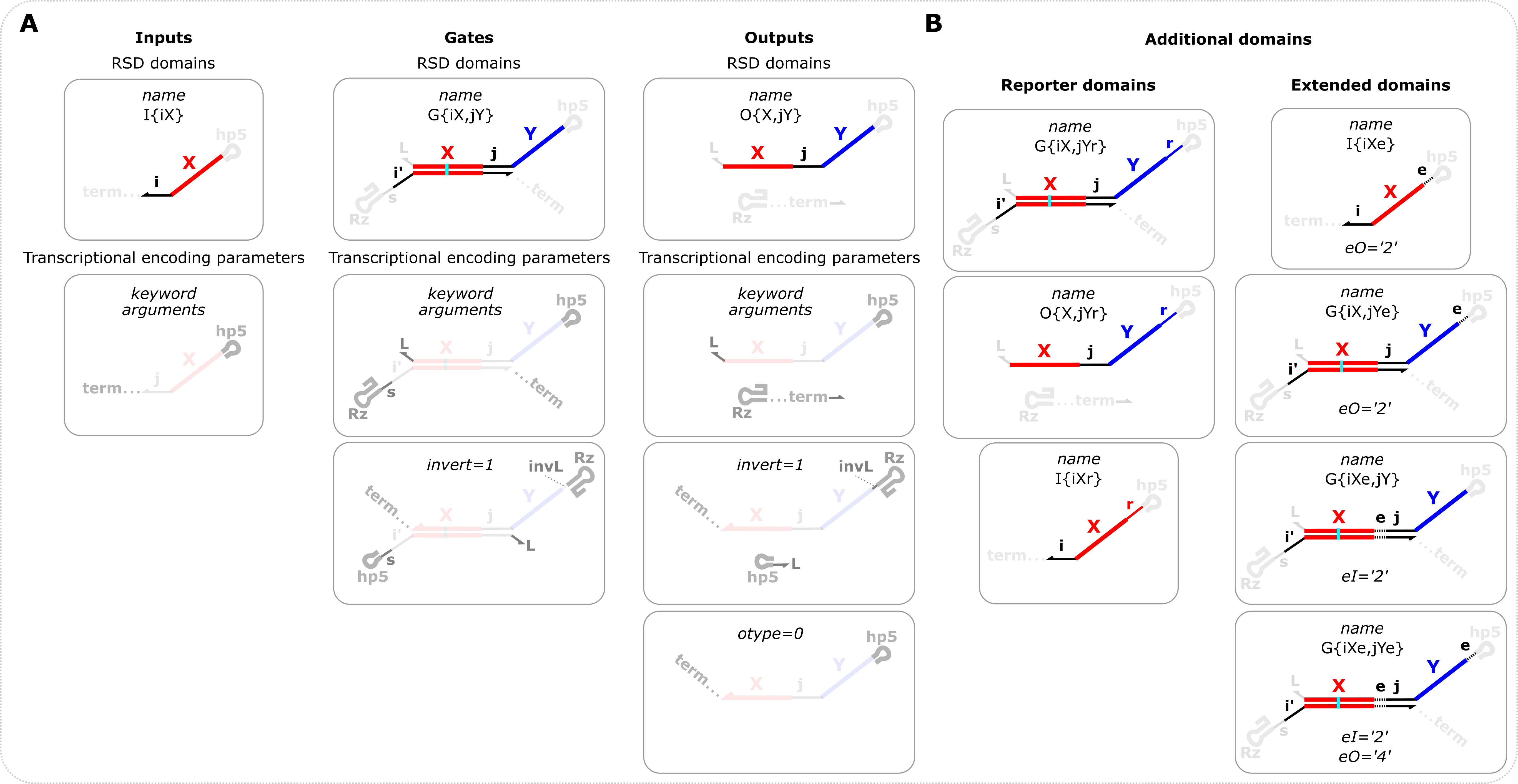
Overview of input, gate, and output definitions and options (A) RSD domains are domains relevant to RNA strand displacement and are specified in the name variable. i,j,k refer to toehold domains and X and Y refer to branch migration domains which can be 1,2,3,4,5,…etc. If only the name variable is specified, the default transcriptional encoding parameters will be used. The transcriptional encoding parameters refer to domains appended to the RSD domains to facilitate transcription of the ctRSD components. The names of the optional keyword arguments are shown for each component in the second row. The third row shows the schematics if the transcription order is inverted by setting the invert keyword argument to 1. For outputs it is also possible to specify otype=0 to produce an output sequence that does not contain a ribozyme. (B) In addition to the options shown in panel A, there are other domains that can be specified. (Left) Reporter domains: domains necessary for irreversible reactions with a fluorescent reporter complex (r). These are specified solely in the name variable. Note there is a unique r domain for each output domain, i.e., an r for domain 1 and an r for domain 2, etc. (Right) Extended domains: domains that extend the branch migration length of components. These can be specified in either the input, output or both domains of a gate. Although not shown, extended domains are also an option for outputs. The existence of an extended domain is specified in the name variable and the identity of the extended domain is specified with the keyword arguments eI and eO. Note that inputs require the eO keyword be used to specify the identity of the output domain.
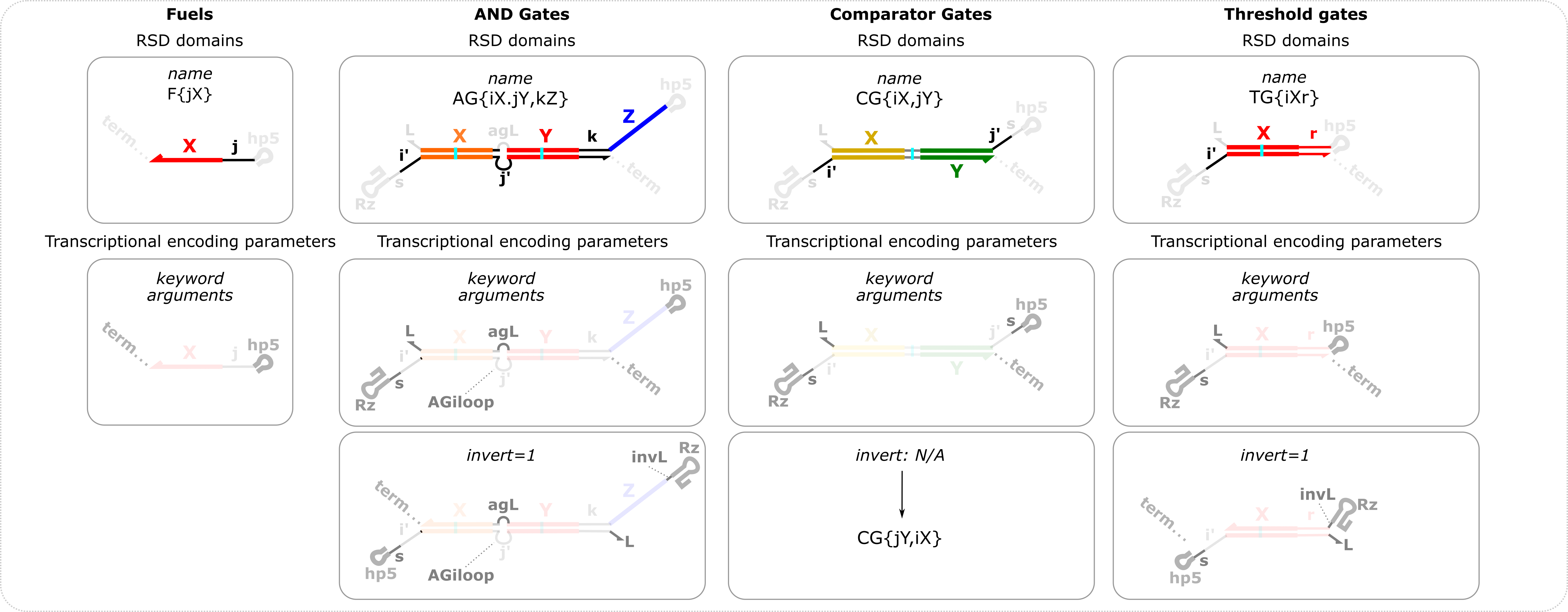
Overview of fuel, AND gate, comparator gate, and thresholding gate definitions and options RSD domains are domains relevant to RNA strand displacement and are specified in the name variable. i,j,k refer to toehold domains and X,Y,Z refer to branch migration domains which can be 1,2,3,4,5,…etc. If only the name variable is specified, the default transcriptional encoding parameters will be used. The transcriptional encoding parameters refer to domains appended to the RSD domains to facilitate transcription of the ctRSD components. The names of the optional keyword arguments are shown for each component in the second row. The third row shows the schematics if the transcription order is inverted by setting the invert keyword argument to 1. Reporter domains can also be specified for AND gates e.g., AG{iX.jY,kZr}. Extended domains are options for fuel and AND gate. For fuel components the convention follows that of input components in the figure above. For AND gates if an e domain is specified for either input domain then that domain will be used for both input domains. But eO can be different than eI for AND gates. Extended domains are currently not an option for threshold gates.
Function Documentation
Warning!
Before all available functions can be accessed the model must be downloaded, imported, and instantiated!
More information on these steps can be found here.
model.ctRSD_seq_compile (name, filepath, Rz=’Ro’, L=’L’, term=’T7t’, hp5=’5hp’, prom=’T7p’, eI=’’, eO=’’, s=’’, invert=0, invL=’A’, agL=’TA’, AGiloop=5, otype=1, rna=0, us=[], ds=[], temp_len=0):
name inputs that show multiple options function with each of those options. All name inputs are also not case sensitive.
- Parameters:
- name: string
- Name of species sequence will be compiled for.
Input (I) -> I{domain} / IN{domain} / INP{domain} / INPUT{domain}
Output (O) -> O{domainI,domainO} / OUT{domainI,domainO} / OUTPUT{domain}
Gate (G) -> G{domainI,domainO} / GATE{domainI,domainO}
AND Gate (AG) -> AG{domainI1.domainI2,domainO} / G{domainI1.domainI2,domainO} / GATE{domainI1.domainI2,domainO}
Fuel (F) -> F{domain}
Threshold gate (TG) -> TG{domain} / T{domains} / TH{domain}
Comparator Gate (CG) -> CG{domainI1,domainI2}
- filepath: string
Local file path for ctRSD domains list Excel sheet. Donwload here. This needs to include the filename of the Excel sheet: /local file path/ctRSD_domains_list.xls
- Rz: string, optional, if NONE,default=’Ro’
Ribozyme sequence. Denoting an ‘x’ before the ribozyme sequence name (for example: ‘xRo’) will use an inactive ribozyme mutant.
- L: string, optional, if NONE,default=’L’
Linker sequence adjacent to the 5’ end of the ribozyme.
- term: string, optional, if NONE,default=’T7t’
Terminator sequence.
- hp5: string, optional, if NONE,default=’5hp’
The sequence of the 5’ hairpin on input and output strands
- prom: string, optional, if NONE,default=’T7p’
Promoter sequence.
- eI: string, optional, if NONE,default=’’
An extended sequence at the 5’ end of output domains on gates, AND gates, outputs, inputs, and fuels. ‘e’ must be specified in the output domain of the name of the species for this input to be valid, i.e., G{u1e,_} or AG{u3e.u1e,_} or O{u5e,_}. eI will be the same for both input domains of an AG.
- eO: string, optional, if NONE,default=’’
An extended sequence at the 5’ end of input domains on gates, AND gates, and outputs. ‘e’ must be specified in the input domain of the name of the species for this input to be valid, i.e., G{_,v1e} or AG{_,w2e} or O{_,v3e} or I{u1e} or F{w5e}. Note that eO is used to specify ‘e’ domains on single input
- s: string, optional, if NONE,default=’’
Spacer sequence between the ribozyme and the input toehold of a gate. Cannot be specified for the second input toehold of an AG. ‘s4’ is the default spacer domain for TG.
- invert: Boolean, optional, if NONE,0
Inverted transcription order - change to 1. This will start transcription at the 5’ end of the input toehold of a gate. Not a valid input for CGs.
- invL: string, optional, if NONE,’A’
For inverted gates only, a linker sequence between the ribozyme domain of the gate and the output domain. Defined as a direct sequence so the default is an ‘A’ base. Any sequence can be directly specified as an input.
- agL: string, optional, if NONE,’TA’
For AND gates only, a linker sequence between the two input domains of an AND gate. Defined as a direct sequence so the default is an ‘A’ base. Any sequence can be directly specified as an input.
- AGiloop: int, optional, if NONE,5
For AND gates only, the number of bases in the internal loop toehold for the second input on an AND gate. This can be 5 bases (default) or 6 bases.
- otype: Boolean, optional, if NONE,1
Specifying the type of output strand to encode. 1 (default) refers to an output that has a ribozyme sequence at the 3’ end to mimic the cleaved output of a ctRSD reaction. 0 refers to an output sequence that ends in a terminator and does not use a ribozyme.
- rna: Boolean, optional, if NONE,0
Set rna = 1 to have the output sequence be the RNA encoded by the DNA template rather than the DNA template sequence. For RNA sequences with a ribozyme a ‘|’ will denote the cleavage site. xRz sequences (inactive ribozyme mutants) will not indicate the cleavage site.
- us: list, optional, if NONE,[]
List of upstream sequences to append to the DNA template. Sequences will be appended 5’ to 3’ upstream of the promoter in the order they are specified in the list.
- ds: list, optional, if NONE,[]
List of downstream sequences to append to the DNA template. Sequences will be appended 5’ to 3’ downstream of the terminator in the order they are specified in the list. The option ‘exc’ can be used in conjtion with temp_length below to create sequences of a specific length.
- temp_len: int, optional, if NONE,0
Specifying the total length of the DNA template. Typically, this can be used to get a template that is 125 bases or 300 bases for ordering as a gBlock or eBlock, respectively. This input should be used in conjunction with us and/or ds to specify which upstream and downstream sequences should be used to meet the length requirement. The option ‘exc’ in ds has a long sequence of bases for appending.
Examples
- First Steps:
Use the example below for guidance
# importing simulator
import sys
sys.path.insert(1,'filepath to simulator location on local computer')
import ctRSD_simulator_200 as RSDs # import latest version of the simulator
# create the model instance
model = RSDs.RSD_sim() # default # of domains (5 domains)
filepath = '//File Path//ctRSD_domains_list.xls'
# use the experimental nomenclature to specify the sequence you want
Gate_seq = model.ctRSD_seq_compile('G{u1,w2r}',filepath)
Gate_seq = model.ctRSD_seq_compile('G{u1,w2r}',filepath,Rz='R3') # specifying an alternative ribozyme sequence
Input_seq = model.ctRSD_seq_compile('I{u1}',filepath)
Fuel_seq = model.ctRSD_seq_compile('F{w1}',filepath)
AG_seq = model.ctRSD_seq_compile('AG{u3.u1,w2r}',filepath)
Example Script for sequences in the 2022 Science Advances paper can be found here
Example Script for diverse gate sequences can be found here
Example Script for additional component sequences can be found here
Example script saving sequence to an Excel file can be found here